Abstract
Comparative analysis of cytoplasmic organelles in a variety of tumors relative to normal tissues generally reveals a strong diminution in mitochondrial content and in oxidative phosphorylation capacity. However, little is known about what triggers these modifications and whether or not they are physiologically reversible. We hypothesized that energy substrate availability could play an important role in this phenomenon. The physiological effects of a change in substrate availability were examined on a human cancer cell line (HeLa), focusing specifically on its ability to use glycolysis versus oxidative phosphorylation, and the effect that energy substrate type has on mitochondrial composition, structure, and function. Changes in oxidative phosphorylation were measured in vivo by a variety of techniques, including the use of two novel ratiometric green fluorescent protein biosensors, the expression level of oxidative phosphorylation and some glycolytic enzymes were determined by Western blot, mitochondrial DNA content was measured by real-time PCR, and mitochondrial morphology was monitored by both confocal and electron microscopy. Our data show that the defective mitochondrial system described in cancer cells can be dramatically improved by solely changing substrate availability and that HeLa cells can adapt their mitochondrial network structurally and functionally to derive energy by glutaminolysis only. This could also provide an explanation for the enhancement of oxidative phosphorylation capacity observed after tumor regression or removal. Our work demonstrates that the pleomorphic, highly dynamic structure of the mitochondrion can be remodeled to accommodate a change in oxidative phosphorylation activity. We compared our finding on HeLa cells with those for nontransformed fibroblasts to help distinguish the regulatory pathways.
INTRODUCTION
Cancer cells are metabolically adapted for rapid growth and proliferation under conditions of low pH and oxygen tension in which nontransformed cells would grow only poorly or not at all (1). Of particular significance, cancer cells generate energy by glycolysis in strong preference to oxidative phosphorylation (OXPHOS; Refs. 2, 3). Associated with this different use of energy yielding substrate is up-regulation of genes for specific isoforms of glucose transport, glycolytic pathway, and conversion of pyruvate to lactate (4). What happens to mitochondria is less clear. Differences in the number of mitochondria (5), their ultrastructure (6, 7), the content and composition in all OXPHOS complexes (5, 8, 9, 10, 11), respiratory chain activity (11, 12), the expression of oxidative phosphorylation dependent genes (13), and levels of mitochondrial DNA (8) have all been variously reported by different investigators, but no consistent pattern has emerged. When glucose is no longer available, as can occur in solid tumors (14), cancer cells are forced to use alternative energy substrates such as the oxidation of glutamine, a process called glutaminolysis (2). This process requires an active oxidative phosphorylation for ATP production. Here, we have examined the physiological effects of such a change in substrate availability on a human cancer cell line (HeLa), focusing specifically on its ability to use glycolysis versus oxidative phosphorylation, and the effect that energy substrate type has on mitochondrial composition, structure, and function. HeLa cells were chosen because they were used in several of the above-referenced studies of mitochondrial form and function and because they proved to readily generate stable cell lines incorporating green fluorescent protein (GFP) reporters of mitochondrial function that we introduced here for examination of mitochondrial functioning in situ.
Our study is the first comprehensive analysis of the changes that occur in the structure and functioning of mitochondria in a transformed cell line. Thus, both substrate-dependent pH and oxido-reduction (REDOX) potential sensors are used to examine the functional state of OXPHOS in vivo, cell growth, mitochondrial network morphology, and the expression levels of various proteins and mitochondrial DNA (mtDNA) content are all quantitated. Our study reveals a highly refined integrated response to substrate availability in cancer cells. The reversible regulation of OXPHOS capacity we observe not only explains how cancer cells respond to glucose limitation in solid tumors (15) but also explains the recovery of OXPHOS observed after therapeutic tumor regression (15, 16) or removal (17).
MATERIALS AND METHODS
Cell Culture Conditions.
The glucose medium consisted of High Glucose DMEM Life Technologies, Inc. (no. −13000), containing 25 mm glucose, supplemented with 10 mm HEPES, 1 mm sodium pyruvate and 10% FCS (Hyclone). The galactose medium consisted of DMEM deprived of glucose (no. 11966-025; Life Technologies, Inc.), supplemented with 10 mm galactose, 2 mm glutamine (4 mm final), 10 mm HEPES, 1 mm sodium pyruvate, and 10% dialyzed FCS (no. SH30079; Hyclone). HeLa cells were kept in CO2 10% at 37°C. Cell proliferation studies were carried out by plating 1.5 × 104 cells on 150-cm2 dishes (Corning) containing 50 ml of glucose or galactose medium. At daily intervals, cells were harvested by trypsinization and counted.
GFP Biosensors Gene Constructs.
To study mitochondrial function in vivo in a noninvasive manner, we targeted a GFP sensitive to pH (GFP pH) or redox potential (rosGFP1) to the matrix space of HeLa cells. The corresponding plasmids, PRA 304 or PRA 306, were created by introducing respectively the mutations S65T/H148D (18) or C48S/T65S/S147C/Q204C into the plasmid pEGFP-N1 (Clontech) containing the leader sequence of the E1α subunit of pyruvate dehydrogenase (PDH) for mitochondrial matrix targeting (a gift from Ruth M. Brown and Gary K. Brown, University of Oxford, Oxford, United Kingdom). Cells were transfected using FuGENE 6 (Roche Molecular Biochemicals) with a ratio of plasmid DNA (μg) to transfection reagent (μl) of 1:2. Stable lines of HeLa cells expressing GFP pH and rosGFP1 in the mitochondrial matrix were obtained through repeated selection of colonies exhibiting green fluorescence with G418 (Calbiochem).
Ratiometric Measurements of Matrix pH and Redox State.
Fluorescence intensity from cells expressing GFP pH or the rosGFP1 was measured at 511 nm with the excitation wavelength consecutively set at 480 and 400 nm. Steady-state measurements of matrix pH (pHm) were performed in a thermostated chamber at 37°C (Princeton apparatus) on individual HeLa cells grown in 35 mm dishes (60–70% confluency), using a Zeiss UEM fluorescence microscope with a ×40, (1.4 numerical aperture), objective dip in-lens (Ultrafluar, Zeiss). Measurements were performed in PBS containing glucose (10 mm) or galactose (10 mm)/glutamine (4 mm). Fluorescence intensity was recorded using an image intensified charge coupled device digital video camera mounted to the camera eyepiece (Princeton Instruments, Trenton, NJ) and OpenLab system software (Improvision). pH calibration curves were obtained in situ by equilibrating cells in various biological buffers (Sigma) of pH ranging from 5 to 11, in presence of monensin (10 μm) as described in Ref. 19. The fit of the data were performed with the software Kaleidagraph (Synergy Software). Steady-state measurements of matrix redox state were performed on a cell suspension (106 cells/ml) in a fluorescence spectrophotometer (Hitachi F4500) with gentle stirring. Cells were grown to 60–70% confluence on a 150-cm2 tissue culture dish, harvested, and suspended in PBS buffer [(pH 7.4; 10 mm Na/K phosphate, 137 mm NaCl, 2.7 mm KCl, 0.5 mm CaCl2, 0.5 mm MgCl2) supplemented with glucose (10 mm) or galactose (10 mm)/glutamine (4 mm)]. The maxima of reduction and oxidation of rosGFP1 were determined in vivo by adding respectively H2O2 (1 mm) or DTT (10 mm) in the cuvette.
Analysis of Mitochondrial Network Morphology in Living Cells.
For the visualization of mitochondrial network in living cells, we used HeLas transfected with the GFP pH cultured on Petri dishes (Corning) in glucose or galactose medium and directly visualized in medium at 37°C on a confocal microscope (Nikon E600/Bio-Rad radiance 2100MP) with a dip in lens Plan Fluor objective (×60 water; numerical aperture 1.4). The excitation was set at 488 nm (Argon-Ion laser tunable infrared laser), and emission was recorded at 511 nm. Images were acquired using Laserpix 4.0 software (Bio-Rad). Three-dimensional reconstruction (projection) was processed with the Laservox software (Bio-Rad).
Respiration Measurements and Determination of Enzyme Activities.
Endogenous cellular oxygen consumption was monitored on intact cells at 37°C in a 1 ml thermostatically controlled chamber (1.0 × 107 cells/ml/run) equipped with a Clark oxygen electrode (Hansatech Oxygraph system). The respiratory buffer was the glucose or the galactose/glutamine growth medium described in the cell culture section. To inhibit mitochondrial respiration rotenone, 5 μg/ml were added in the cuvette. Cytochrome c oxidase (COX) activity was measured in the Oxygraph after addition of 3 mm ascorbate, 0.5 mm N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD), and 10 μg/ml antimycin. KCN sensitivity of (COX) was verified by adding KCN 20 μm in the cuvette. The endogenous respiratory rate and COX activity were expressed in nmol O2/min/106 cells.
Transmission Electron Microscopy.
Cells grown in glucose or galactose medium were fixed for Spurr’s resin embedment in 2.5% glutaraldehyde, 4% paraformaldehyde in 0.1 m sodium cacodylate (pH 7.4), with 8 mm CaCl2 followed by 1% osmium tetroxide (pH 7.4), followed by ethanol dehydration. Ultrathin sections were counterstained with lead citrate and viewed on a Philips CM12 electron microscope.
Western Blotting.
Cells grown in glucose or galactose medium were washed in PBS, harvested with a cell scraper, frozen overnight, and lysed (PBS, lauryl-maltoside 2%) 10 min on ice. After centrifugation of the cell lysate (300 × g, 10 min), soluble proteins were isolated in the supernatant, and protein concentration was determined according to the BCA method (Pierce). Samples were dissolved in SDS-PAGE tricine sample buffer (Bio-Rad) containing 2% β-mercaptoethanol by incubation for 30 min at 37°C and separated on a 10–22% SDS-polyacrylamide gradient mini-gel. Proteins were transferred electrophoretically to 0.45 μm of polyvinylidine difluoride membranes for 2 h at 100 mAmp in 3-(cyclohexylamino)propanesulfonic acid buffer (3.3 g of 3-(cyclohexylamino)propanesulfonic acid, 1.5 liters 10% methanol (pH 11.00)] on ice. Membranes were blocked overnight in 5% milk-PBS +0.02% azide and incubated for 3 h in a mixture of monoclonal antibodies against respiratory chain complexes (Monoclonal Antibody Facility, University of Oregon, Eugene, OR, also available from Molecular Probes). We also used commercially available antibodies for the detection of Porin (Calbiochem), Tubulin (Molecular Probes), Hexokinase I, II, and Glut 1 (Santa Cruz Biotechnology). For the GFP studies, we used a polyclonal antibody from (Clontech). After three washes with PBS-0.05% Tween 20, the membranes were incubated for 2 h with horseradish peroxidase-conjugated goat antimouse (Bio-Rad) diluted in 5% milk-PBS. This secondary antibody was detected using the chemiluminescent ECL Plus reagent (Amersham).
Quantitative PCR.
Total DNA was extracted from cultured cells using Qiagen DNeasy tissue kit according to manufacturer recommendation (Qiagen). The amount of mtDNA was determined using a Perkin-Elmer 7700 TaqMan PCR machine (Perkin-Elmer, CA) in triplicate. The forward primers were 5′-GCTACCTAAGAACAGCTAAAAGAGC-3′ (1918 to 1942), the reverse primer was 5′-AAGATTCTATCTTGGACAACCAGC3′ (2038 to 2015), and probe was FAM-ATGTAGCAAAATAGTGGGAAGA-TAMRA (2012 to 2033). The mtDNA quantity was corrected by simultaneous measurement of the nuclear DNA, a 28SrRNA. The primers for 28SrRNA were 5′-AACGAGATTCCCACTGTCCC-3′ and 5′-CTTCACCGTGCCAGACTAGAG-3′. The probe was FAM-TATCCAGCGAAACCAC-TAMRA. The amplification conditions were 50 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, with an initial 10 min of extra denaturation at 95°C. The amount of mtDNA was expressed by relative quantity of 28SrRNA using Ct values calculated by the sequence detector v1.6 (Perkin-Elmer).
RESULTS
Cells in Culture Can Derive Energy from OXPHOS When Glucose Is Not Available.
Frozen HeLa cells previously grown in high-glucose medium were thawed and grown for 15 cell divisions on either glucose or galactose/glutamine medium. The medium was replaced every 3 days, and the cells were replated when confluency was reached. Cell growth rates were measured over 10 days. In glucose medium, HeLas divided rapidly with a doubling time of ∼1 day (23.9 ± 1 h). In galactose/glutamine medium, the doubling time was three times longer (58 ± 2 h). The rate of acidification of the medium induced by cell proliferation could be followed by the color change of the pH indicator phenol red. When cells reached confluency without replacing the medium, the pH was more acidic in glucose medium (pH ≈ 6.8) than in galactose where it remained close to the original value (pH ≈ 7.4). This difference can be attributed to a larger production of lactate when these cell lines are grown in glucose medium for even a short time (2). No significant change in cell morphology or cell death rate was observed in HeLas grown in galactose compared with glucose. Culture of a primary (lung) fibroblast cell line (MRC5) was also significantly slower in galactose/glutamine than in glucose (data not shown). However, when allowed to go acid through lactic acid build up, fibroblasts died, in contrast to the prolonged survival of the HeLa cells under the same conditions.
Mitochondrial Respiration Is Stimulated in Galactose Medium.
The rotenone-sensitive endogenous respiratory rate and the cyanide sensitive COX activity of intact HeLa cells grown in galactose or glucose were measured by polarography to assess directly the functional state of mitochondrial oxidative phosphorylation. As shown in Table 1, these activities were significantly increased in the galactose medium, i.e., the endogenous respiratory rate value was 1.35 ± 0.23 nmol/O2/min/106 cells in glucose compared with 2.72 ± 0.36 in galactose, and maximal COX activity value was 5.60 ± 1.38 nmol/O2/min/106 cells in glucose compared with 17.35 ± 5.0 in galactose for HeLa cells.
Steady-State Mitochondrial pHm Is 0.4 Units Lower in Galactose.
Mitochondrial matrix pH was measured in vivo in HeLa cells using a GFP that acts as a pH indicator (Ref. 18; GFP pH-S65T/H148D). This GFP exhibits two pH-dependent excitation peaks with an isobestic point that allows ratiometric pH measurements. HeLa cells were stably transfected with this GFP-pH targeted to the matrix space, as confirmed by fluorescence microscopy and by immunogold labeling in transmission electron microscopy (data not shown). To calibrate the signal in situ, the fluorescence ratio (480/400 nm) was measured in individual cells placed in various biological buffers (pH ranging from 5 to 11) in the presence of 10 μm monensin to allow equilibration of the matrix pH with that of the buffer (19). The calibration curves obtained on individual HeLa are shown in Fig. 1,A. The sigmoidal shape describes an acid-base equilibrium for a single titratable group as expected (18). The fit of the data to a pH titration curve gave a pKa value of 8.51 ± 0.07 for the GFP pH in HeLa cells. In vivo titration curves were used to determine the steady-state pH value of the mitochondrial matrix (pHm). The matrix pH was lower in the galactose medium (Table 1), e.g., pH 8.03 ± 0.05 in glucose compared with 7.65 ± 0.10 in galactose. Finally, the uncoupler carbonyl cyanide m-chlorophenylhydrazone was used to verify that GFP-pH responded to the dissipation of the pH gradient across mitochondrial inner membrane. Addition of 10 μm carbonyl cyanide m-chlorophenylhydrazone to HeLa cells grown in either glucose or galactose always resulted in a strong acidification of the matrix down to a pHm value close to 7 (Table 1).
Matrix Redox State Is More Oxidized in Galactose Than Glucose.
As another way to evaluate the steady-state level of oxidative phosphorylation in living cells, the mitochondrial matrix redox state was measured in vivo, using a GFP that is a ratiometric redox potential indicator (rosGFP1; Ref. 20). This redox-sensitive GFP was created by directed substitution of cysteine residues at selected surface sites in the vicinity of the GFP chromophore. A similar GFP redox sensor, although nonratiometric, was described by Ostergaard et al. (21). Data for HeLa cells are presented in Fig. 1,B. After the addition of 1 mm H2O2, the rosGFP1 intensity ratio (480/400 nm) was set as the 0% reduction, and the 100% reduction was consecutively obtained by an addition of 10 mm DTT (Fig. 1,B). Both effects were reversible. With respect to this scale, the relative measurements of matrix redox potential were 93 ± 3% reduction of rosGFP1 in cells grown in glucose medium compared with 74 ± 2% in galactose medium (Table 1).
HeLa Cells Synthesize More Respiratory Chain Protein When Forced from Glycolysis to Growth by OXPHOS.
Expression levels of various mitochondrial proteins involved in energy metabolism were examined by quantitative Western blot for HeLa cells grown for 15 doublings in glucose or galactose medium. Monoclonal antibodies directed against subunits of respiratory chain complexes I-V, along with PDH E1α subunit, citrate synthase (Krebs cycle), and porin were used for this analysis (Fig. 2). The quantitation of OXPHOS complexes with this set of monoclonal antibodies has been reported previously for fibroblast cell lines with different mitochondrial defects (22, 23). The levels of the different complexes were determined under conditions where the signal from bound antibodies was not saturated (Fig. 2,A), and the data are expressed as the protein band intensity value (Fig. 2 B). Equivalent loading of protein samples was verified by Sypro-Ruby (Molecular Probes, Inc.) staining of the gel or by using tubulin as a standard protein control. The results show a significant increase (factor of 2) in the expression level of porin, citrate synthase, respiratory chain complexes I, II, V, PDH, and a stronger increase (factor of 3) in subunit 5a of COX/μg cell protein in HeLa cells grown in galactose for 3 weeks. Only complex III of the OXPHOS components was not significantly altered by substrate conditions.
To see whether the increased expression of OXPHOS components observed in HeLa cells grown in galactose occurred in nontransformed cells, similar experiments were conducted on primary fibroblasts grown for 3 weeks in glucose or galactose medium. The results (Fig. 3) show a significant increase in the expression of CIV (5a), CI (20 kDa), Porin, CVα and CII (70 kDa) in galactose-grown MRC5s, although less than the 2- or 3-fold seen in HeLa cells. No increase was detected for CIII (core 2) or for PDH (E1α).
Glycolysis Enzymes Remain High Based on Hexokinase and Glucose Transporter.
To see whether growing HeLa cells in the absence of glucose for 15 doublings could affect the high levels of glycolytic enzymes, we measured the expression levels of particular proteins involved in glucose uptake and metabolism in HeLa cells grown in glucose or galactose medium. For that, we performed Western blotting using polyclonal antibodies against Hexokinase isoforms I and II and against glucose transporter Glut 1 (Fig. 4). The results show no significant difference in the expression levels of Hexokinase I, II, and Glut 1 in HeLa cells grown in glucose or in galactose for three weeks (Fig. 4).
mtDNA Content Is Unchanged in HeLa Cells Grown on Galactose.
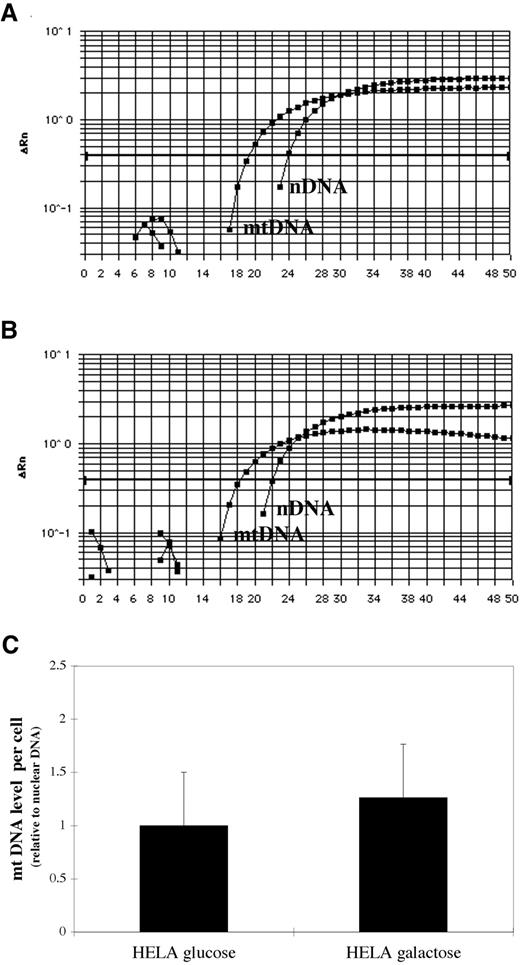
mtDNA content in HeLa cells grown in glucose or galactose medium was measured by real time PCR to determine whether forcing cells to derive energy by OXPHOS had an effect on mitochondrial genome replication (24). To correct for quantitative variations among the samples such as differences in cell number, mtDNA content was determined simultaneously with nuclear DNA content (Fig. 5, A and B), and mtDNA levels were normalized relative to nuclear DNA signals (Fig. 5 B). The data reveal no significant difference in mtDNA content/cell between HeLas grown in glucose (1.000 ± 0.6134) or in galactose (1.263 ± 0.962) for 3 weeks (P = 0.7103, unpaired two-tailed Student’s t test).
Mitochondria of Cells Grown in Galactose Show the Condensed Configuration.
The differences in mitochondrial structure that result from growing HeLa cells in glucose compared with galactose were examined by electron microscopy. When compared with cells grown in high glucose medium (Fig. 6, A and B), galactose-grown cells (Fig. 6, D and E) showed consistent increase in matrix density due to condensation of the mitochondrial matrix and expansion of the cristal spaces, i.e., the classical orthodox to condensed transformation, which occurs with activation of OXPHOS as described earlier by Hackenbrock et al. (25). As a result, at low magnification and using the staining procedures commonly used for electron-microscopy (EM) analysis, mitochondria were difficult to find in the cytosol of cells grown in glucose medium (Fig. 6,A), whereas in galactose, they were more clearly apparent (Fig. 6,D). Electron microscopy of nontransformed fibroblasts cultured in the different energy substrates also showed differences in mitochondrial morphology (Fig. 6, C and F). Almost all of these organelles were in the condensed configuration after growth on galactose/glutamine (Fig. 6,F), whereas only a small fraction were in this state in cells grown on glucose (Fig. 6 C).
Cells Grown in Galactose Have More Cristae but not More Mitochondrial Mass.
Visualization of the internal organization of mitochondria in glucose- and galactose-grown HeLa cells illustrates the substrate-dependent differences in mitochondrial morphology. Image analysis of high magnification transmission electron microscopy micrographs (10 for each cell culture conditions) allowed us to quantify the internal compartments and membranes of several mitochondrial sections (100 different sections) in cells grown in different substrates (Table 2). When compared with cells grown in high glucose medium (Fig. 6,B), galactose-grown cells (Fig. 6,E) showed consistent increases in the amount of cristal membranes (Table 2). Additional cristae were not present in fibroblasts grown on galactose/glutamine (Fig. 6,F) compared with glucose (Fig. 6 C) or were not detected because the effect was much less dramatic than for HeLa cells (results not shown).
The results summarized in Table 2 show that there was no statistical increase in the number of mitochondrial sections in HeLa cells forced to use OXPHOS to derive energy. For instance, the number of mitochondrial sections/cell was (19.8 ± 7.3) in galactose compared with (15.6 ± 6.8) in glucose. Mitochondrial area/cell area was 5.4 ± 0.94 in glucose compared with 4.2 ± 1.96 in galactose. Also, there was no significant difference in the number of mitochondrial sections of fibroblasts grown under the two substrate conditions.
Alterations in the Mitochondrial Reticulum in Galactose.
To examine the reticulum arrangement, cells were examined by fluorescence confocal microscopy using the matrix-targeted GFP-pH as the mitochondrial marker (Fig. 7). In HeLa cells grown in glucose medium, the mitochondrion appeared as a reticulum mostly clustered in the perinuclear region and not typically extending far out into the cell (Fig. 7,A). By contrast, in galactose-grown cells, the mitochondrial reticulum extended outward much more into the additional processes of the cell (Fig. 7,C) and appeared to be more elaborately interconnected and ramified. The same differences were not observed for fibroblast cells (Figs. 7, B and D). In addition, the mitochondrial reticulum was made up of narrower tubules in galactose-grown cells than in glucose grown cells, 0.48 versus 0.32 μm in HeLa cells (Table 2). In fibroblasts, the thinning of the reticulum was not as dramatic and statistically not significant (Fig. 7 D).
Galactose-grown cells frequently displayed rings joined with one another by short filaments (Fig. 7,C). Although such rings are seen in cells grown in glucose, they were very much less frequent. These rings were not observed in galactose grown MRC5 fibroblasts. Consistent with the restructuring of the mitochondrial reticulum, EM of galactose-grown HeLa cells showed more heterogeneity of the size of mitochondrial profile than glucose-grown cells because of sectioning through long stretches of reticulum, as well as heavily looped regions (Fig. 6,D). Finally, there were many more electron-dense inclusions in the matrix space of HeLa galactose-grown than glucose-grown cells (Table 2). For instance, we observed only one inclusion/cell in glucose, whereas in galactose medium, the average number of inclusion bodies/cell was 25. This difference was not observed in fibroblasts.
Time-Dependent Analysis of Mitochondrial Restructuring.
To know how soon after the change of energy substrate the modifications in mitochondrial morphology become detectable, we performed a time-dependent analysis of mitochondrial structure on HeLa cells grown in glucose, rapidly washed in PBS, and then switched to galactose. After the first hour of growth in galactose medium, no difference could be observed by EM or fluorescence microscopy (data not shown). After 1 day, by which time most of the glucose endogenous to the cells had been used, we observed matrix condensation in 60% of the cells; after a week, all of the cells showed condensed mitochondria. The significant changes in the ramification and interconnection of the mitochondrial network were only observed after 2 weeks of galactose treatment. Thus, the condensation of the mitochondrion as cells begin to use glutamine precedes interconnection and ramification of the network.
DISCUSSION
Cells must respond to changes in environment, including availability of substrates for energy metabolism, if they are to survive. Here, we have examined the adaptations of a transformed cell line, HeLas, and a primary cell line, fibroblasts, when forced to change from generating ATP predominantly by glycolysis to exclusively by oxidative phosphorylation. HeLas, as cancer cells, are specialized to generate their energy by glycolysis, probably to survive under conditions of hypoxia where oxidative phosphorylation is not possible. Extensive glycolysis produces lactic acid, which makes the surrounding medium acid. Cancer cells can survive in an acid medium, which is detrimental to surrounding noncancer cells, and this improves the invasiveness of a tumor. It has been shown that when HeLa cells are grown in glucose medium, ∼80% of the glucose is metabolized through glycolysis to lactic acid and only 4–5% of sugar carbon entered the Krebs cycle (2). In contrast, when HeLa cells were grown in galactose medium, glutamine provided 98% of the ATP used for growth (2). Fibroblasts, a nontransformed cell line, also use glycolysis in tissue culture when glucose levels are high but retain significant levels of oxidative phosphorylation at the same time (2, 26). Here, we show that both cell types can survive when making ATP exclusively by oxidative phosphorylation and that HeLa cells do so by remodeling their mitochondria. We observed similar changes in another transformed cell line, 143B osteosarcoma (data not shown), suggesting that substrate-driven mitochondrial changes could be a general phenomenon in cancer cells. Although the change is much more dramatic in HeLas and osteosarcomas, fibroblasts appear to make some of the same structural and functional changes observed in HeLas, so the pathways used could be general and not unique to transformed cells.
There was a 2-fold increase in endogenous respiratory rate of HeLa cells in galactose medium versus in glucose medium. Possible explanations are a general stimulation of mitochondrial biogenesis, an increased synthesis of respiration-related enzymes, an increased flux of glutamine oxidation through an existing pathway (Krebs-cycle and respiratory chain), a transition from state 4 to state 3, or a combination of each. To investigate actual respiratory pathway activation in HeLa cells grown in galactose medium, we assessed the status of oxidative phosphorylation in vivo with GFP biosensors targeted inside the mitochondria. Both the steady-state pH and redox state inside mitochondria were measured using two novel GFP biosensors that are ratiometric and which should find widespread use in mitochondrial studies. The ratiometric GFP pH (18) and rosGFP1 (20) probes allow determination of pH or redox potential within cells or between groups of cells, independent of photobleaching, indicator concentration, variable cell thickness, illumination stability, excitation path length, and nonuniform indicator distribution (27). In addition, these two ratiometric dual-excitation GFP biosensors present a high fluorescence, protease resistance, and stability throughout a wide range of pH and solvent conditions.
HeLa cells stably transfected with the GFP-pH (targeted to the matrix space) revealed a more acidic pH inside the mitochondria of cells grown in galactose medium compared with glucose. This is in agreement with the work of Llopis et al. (19) who measured a pHm value of 7.98 ± 0.07 in HeLa cells grown in 10 mm glucose medium, and a value ∼7.4 after the addition of an oxidizable substrate (pyruvate/lactate), whereas a study performed in 293T and Jurkat cell lines (28) gave pHm value of 8.1 and 7.8, respectively. In contrast to our work, both of these previous studies used a nonratiometric GFP-pH. As pointed out by Takahashi et al. (29), dual-excitation ratiometry not only increases the dynamic range of pH measurements but also corrects for differences in the amount of expression of the biosensor within and between different cells.
The matrix space was more acid by 0.4 units in cells grown on glutamine than on glucose. OXPHOS generates an electrochemical gradient of protons that consists of both a pH gradient (ΔpH) and a membrane electric potential (ΔΨ). Several bioenergetic studies of oxidative phosphorylation functioning in isolated mitochondria have measured the transition from resting (slow respiring) state 4 to phosphorylating (fast respiring) state 3 and find that this leads to a decrease in both ΔΨ and in ΔpH (e.g., Ref. 30). In isolated rat liver mitochondria, the ΔΨ decreases from approximately −230 to −170 mV (31) and the ΔpH from ∼0.8 to 0.4 (32), the same ΔpH we measure in vivo. This is consistent with cells grown in galactose being in state 3, whereas those in glucose are in state 4. The acidification of pHm we and others (33) observe at state 3 is mostly due to the activity of various transporters that use the ΔpH to deliver essential metabolites inside the mitochondria for oxidative phosphorylation, e.g., the entry of inorganic phosphate (Pi) via the Pi−/OH− exchanger.
In addition to substrate effect on steady-state pH, the GFP-redox state indicator (rosGFP1) showed a more oxidized matrix redox state in the galactose medium. Our studies on rosGFP1 have indicated that NADH is the most likely reductant of the probe in the mitochondrial matrix (20) and that this reduction is mediated by enzymes such as PDH or peroxiredoxin (34), which are two-electron acceptors but one-electron donors. Thus, the different reduction levels of rosGFP1 in glucose or galactose medium are probably because of a difference in the NADH/NAD+ ratio inside the mitochondria. This ratio has been evaluated in fibroblast cell cultures by measuring the lactate/pyruvate ratio in the medium and estimated to be 6:1 in the presence of glucose compared with 1:10 in absence of glucose (35). The highly reduced matrix redox state observed here in the presence of glucose is a reflection that the mitochondria are unable to efficiently reoxidize the pyruvate or the NADH produced by glycolysis and correlates with the important acidification of the medium observed in HeLa cells grown in glucose.
According to John (36), tumor-derived cells cultured in glucose medium can produce 40 times more lactic acid than normal cells. This reaction can only occur when the cytosolic NADH/NAD+ ratio is high as we observe here, and/or when the pyruvate (normal end product of glycolysis) does not enter the mitochondria for complete oxidation by the PDH, the Krebs cycle and mitochondrial oxidative phosphorylation. The molecular bases of this phenomenon are not clearly understood. Several suggestions have been proposed including a reversion to the fetal program of gene expression (37), mutations in mtDNA (38), and excessive ROS production (39). Pedersen (4) has also proposed that tumor-derived cells up-regulate glycolytic enzymes by increased protein synthesis and by switching isozyme types in favor of species with reduced rates of back reactions. They also have an increased binding of Hexokinase (isoform 2) to the mitochondrial outer membrane, which reduces Pi and ADP delivery to the OXPHOS system (40). At the same time, they down-regulate mitochondrial functioning by altering amounts and throughput of substrates in the organelle. In addition to all of these modifications linked to the transformation process, our results show that cancer cells metabolic profile can also be modified by solely changing substrate availability for several doublings without altering the metabolic demand of tumor cells per se (41).
Taken together, the various live cell functional measurements demonstrate a much more active state of oxidative phosphorylation in cells grown in galactose than those in glucose medium do.
Energy Substrate-Dependent Changes in Mitochondrial Composition.
There are increased levels of most of the proteins of the respiratory chain as well as the ATP synthase/mg of cell protein in HeLas and fibroblasts when grown in galactose plus glutamine, compared with glucose. Complexes I, II, IV, and V are all increased in amount by at least 2-fold in HeLas and significantly, although <2-fold, in fibroblasts. Interestingly, the levels of complex III were not increased concomitantly, which suggests that higher amounts of this complex are needed even when there is little or no OXPHOS, possibly for other electron transfer reactions or because of biosynthetic needs. One interesting difference between the transformed and nontransformed cell line is the change in levels of PDH complex. In HeLa cells, the levels of this enzyme are highly up-regulated in OXPHOS, which this is not the case in fibroblasts. It may be that down-regulation of the levels of PDH is a specialized feature of cancer cells that is programmed along with up-regulation of Glut4 and the switch of glycolytic enzyme isoforms. In HeLa cells, there was also an increase in citrate synthase protein expression (2-fold), which could indicate induced expression of Krebs cycle enzymes, although expression levels were not examined directly. This can be related to the fact that in galactose medium energy for growth is derived via glutamine, which is catabolized by mitochondrial glutamase into glutamate, that is then converted into α-ketoglutarate by glutamate dehydrogenase and additionally processed in the Krebs cycle.
Quantification of mtDNA content by real-time PCR showed no difference in HeLa cells grown in glucose or in galactose media. Therefore, the observed increase in OXPHOS proteins in HeLa cells grown in galactose is the result of a regulation of OXPHOS genes expression rather than an augmentation of mtDNA content. The possibility that the observed increase in OXPHOS proteins is caused by a clonal selection of the cells that contained originally the highest levels of OXPHOS components is unlikely because there was no significant increase in the number of dead cells in galactose medium.
Energy Substrate-Dependent Changes in Mitochondrial Structure.
Observation of the mitochondrial network in living cells has been made possible by the use of fluorescent probes that accumulate as a function of the transmembrane electric potential (ΔΨ; Ref. 42) or by the use of GFPs targeted to the matrix space (43). Recent observations of the internal structure of mitochondria by electron tomography have revealed six discrete compartments: outer membrane; inner membrane; cristal membrane; intracristal space; intermembrane space; and matrix (44). Our analysis of mitochondrial morphology using both fluorescence and electron microscopy reveals important transformations of mitochondrial overall structure and internal organization in response to a change in metabolic substrate that have not been seen previously. There is considerable structural remodeling to allow and accommodate the switch from glycolysis to oxidative phosphorylation. The additional amounts of OXPHOS components observed in HeLa cells grown on galactose are accommodated in mitochondria by increasing the amounts of cristal membrane rather than any major increase in the mass of mitochondria as shown by the microscopy data and by the observation that there is no significant increase in the levels of mtDNA, which would be expected for increased mitochondrial mass. In galactose grown HeLa cells, we observed a 3-fold increase in the ratio of cristal membrane to inner membrane that we think could be related to the 2-fold increase in respiratory chain complexes synthesis. Indeed, we have already shown that in rho° cells (devoid of mtDNA), the inner membrane surface is reduced (45), and our recent data demonstrate that mitochondrial respiratory complexes are preferentially localized to the cristal membrane (46), suggesting that the mitochondrial cristae comprise a regulated submitochondrial compartment specialized for ATP production.
When producing energy by oxidative phosphorylation, both HeLa and fibroblast mitochondria adopt the condensed configuration, whereas during glycolysis, they maintain an orthodox state. The switching of mitochondria between these two forms has been described first by Hackenbrock et al. and others (25, 47, 48), but this is the first study to show this switching in vivo. To know how soon after the change of energy substrate the modifications in mitochondrial morphology become detectable, we performed a time-dependent analysis of mitochondrial structure on HeLa cells grown in glucose and then switched to galactose. The results show that we have to distinguish two types of morphological transformations: (a) a rapid condensation of the mitochondrion; and (b) a slow interconnection and ramification of the network. Hackenbrock et al. (25) reported that the orthodox to condensed transition occurred in Ehrlich ascites cells only 6 s after the intracellular generation of a burst of ADP with 2-deoxyglucose. In our work, the longer time necessary to observe such ultrastructural transformation of the mitochondrion could be because OXPHOS activation may have occurred only when all of the glucose already present in the cells was metabolized. Our results also differ from the work of Hackenbrock et al. (25) in the manner used to stimulate OXPHOS; we induced a steady-state activation of OXPHOS by physiological substrate utilization instead of generating rapidly a burst of ADP in the cell.
Along with switching to a condensed state, the mitochondria of cells adapted to making ATP by OXPHOS are in a considerably more complex reticular form. This reticulum extends throughout the cell rather than being perinuclear as in glucose-grown cells. It thins and becomes much more looped as shown most clearly for HeLa cells. The ramification of the reticulum for OXPHOS may be important in generating ATP at all parts of the cell. In glycolysis, the mitochondria may cluster at the nucleus for efficiency of their now predominantly biosynthetic functions. The two processes, the orthodox-to-condensed change and the thinning and ramification of the reticulum, are not linked. The orthodox-to-condensed switch occurs as a substrate is changed, whereas the thinning and ramification occur late and presumably result from increased OXPHOS protein synthesis. This mitochondrial remodeling observed in HeLa cells is different from the adaptation of OXPHOS mitochondrial mutants that generally increase total mitochondrial mass (49). When forced to use OXPHOS, cancer cells that also present a deficient mitochondrial system compared with their tissue of origin (4, 8) could have been expected to lead to the same adaptative modifications of the mitochondrion. On the contrary, it is the condensation, thinning, and ramification of the mitochondrial reticulum that dominates the changes.
In summary, cells such as HeLas, osteosarcomas 143B, and fibroblasts can live using glycolysis or oxidative phosphorylation, but HeLa and osteosarcoma cells do so by altering significantly mitochondrial composition and form to facilitate optimal use of the available substrate. The signaling pathways by which this is accomplished must include transcription factors that alter expression of OXPHOS components. This increased protein synthesis starts a complicated process of structural remodeling, the mechanism of which remains to be solved. However, such cells in culture may not resemble a real tumor, and the mitochondrial adaptation that we describe could be different in vivo and vary with the tissue. The factors determining mitochondrial structure in tissues could be substrate nature and availability, oxygen partial pressure, energy demand, and mitochondrial respiratory steady state. In our study, we solely analyzed the effect of a change in substrate nature on mitochondrial composition. Mitochondrial network remodeling could be of particular importance for the survival of large tumors (14). Understanding the molecular mechanisms that force the cell to develop its mitochondrial respiratory capacity in galactose medium should provide insight into the regulation of mitochondrial biogenesis, as well as in the formation of the tumor metabolome (50), which in turn could lead to new targets for the gene therapy of cancer and also mitochondrial disorders (51).
Grant support: R. Gilkerson was supported by NIH Training Grant GM07759.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Note: R. Rossignol and R. Gilkerson contributed equally to this work.
Requests for reprints: Rodrigue Rossignol. Phone: 33-5-57-57-16-57; Fax: 33-5-57-57-16-12; E-mail: rossig@u-bordeaux2.fr
Live cell measurements of mitochondrial matrix pH and redox state using green fluorescent (GFP) biosensors. A, titration curve of matrix targeted GFP pH in HeLa cells. Cells were incubated in various calibration buffers (pH 5–11) at 37°C in presence of monensin (10 μm). Fluorescence intensity was recorded at 511 nm with excitation set at 400 and 480 nm. Ratio of fluorescence intensity excited at 480 nm to that at 400 nm is plotted as a function of pH. B, fluorescence excitation spectra of HeLa cells transfected with rosGFP1 after 1 mm H2O2 addition (———) and after 10 mm DTT addition (· · ·). Steady-state measurements of matrix redox state were performed on a cell suspension (106 cells/ml) in a fluorescence spectrophotometer (Hitachi F4500) with gentle stirring. Spectra were collected while monitoring emission at 510 nm.
Live cell measurements of mitochondrial matrix pH and redox state using green fluorescent (GFP) biosensors. A, titration curve of matrix targeted GFP pH in HeLa cells. Cells were incubated in various calibration buffers (pH 5–11) at 37°C in presence of monensin (10 μm). Fluorescence intensity was recorded at 511 nm with excitation set at 400 and 480 nm. Ratio of fluorescence intensity excited at 480 nm to that at 400 nm is plotted as a function of pH. B, fluorescence excitation spectra of HeLa cells transfected with rosGFP1 after 1 mm H2O2 addition (———) and after 10 mm DTT addition (· · ·). Steady-state measurements of matrix redox state were performed on a cell suspension (106 cells/ml) in a fluorescence spectrophotometer (Hitachi F4500) with gentle stirring. Spectra were collected while monitoring emission at 510 nm.
Mitochondrial protein expression levels are increased significantly in galactose compared with glucose-grown HeLa cells. A, densitogram representing the expression levels of complex IV subunit 5a (COX 5a), complex I subunits 20 kDa (CI 20kD) and 30 kDa (CI 30kD), porin, pyruvate dehydrogenase subunit E1α (PDH), complex III subunit (core 2), F1F0 ATPase subunit α (CV α), complex II 70 kDa (CII 70kD), and citrate synthase (CS) examined by quantitative Western blot using monoclonal antibodies developed in our laboratory. B, the levels of the different complexes obtained for glucose grown ( ) or galactose grown (
) or galactose grown ( ) HeLa cells were determined by densitometry under conditions where the signal from bound antibodies was not saturated. The data are expressed as the protein band intensity value n ≥ 5, ∗, P ≤ 0.05 galactose versus glucose.
) HeLa cells were determined by densitometry under conditions where the signal from bound antibodies was not saturated. The data are expressed as the protein band intensity value n ≥ 5, ∗, P ≤ 0.05 galactose versus glucose.
Mitochondrial protein expression levels are increased significantly in galactose compared with glucose-grown HeLa cells. A, densitogram representing the expression levels of complex IV subunit 5a (COX 5a), complex I subunits 20 kDa (CI 20kD) and 30 kDa (CI 30kD), porin, pyruvate dehydrogenase subunit E1α (PDH), complex III subunit (core 2), F1F0 ATPase subunit α (CV α), complex II 70 kDa (CII 70kD), and citrate synthase (CS) examined by quantitative Western blot using monoclonal antibodies developed in our laboratory. B, the levels of the different complexes obtained for glucose grown ( ) or galactose grown (
) or galactose grown ( ) HeLa cells were determined by densitometry under conditions where the signal from bound antibodies was not saturated. The data are expressed as the protein band intensity value n ≥ 5, ∗, P ≤ 0.05 galactose versus glucose.
) HeLa cells were determined by densitometry under conditions where the signal from bound antibodies was not saturated. The data are expressed as the protein band intensity value n ≥ 5, ∗, P ≤ 0.05 galactose versus glucose.
Nontransformed MRC5 fibroblasts grown in galactose for 15 doublings show an increased expression of some OXPHOS proteins. Expression levels of complex IV subunit 5a (COX 5a), complex I subunits 20 kDa (CI 20kD), porin, pyruvate dehydrogenase subunit E1α (PDH), complex III subunit (core 2), F1F0 ATPase subunit α (CV α), and complex II 70 kDa (CII 70kD) were examined by quantitative Western blot using monoclonal antibodies developed in our laboratory on glucose grown ( ) or galactose grown (
) or galactose grown ( ) MRC5 fibroblasts. The levels of the different complexes were determined by densitometry under conditions where the signal from bound antibodies was not saturated for 5, 10, and 15 μg of proteins. The data are expressed as the protein band intensity value n ≥ 2, ∗, P ≤ 0.05 galactose versus glucose.
) MRC5 fibroblasts. The levels of the different complexes were determined by densitometry under conditions where the signal from bound antibodies was not saturated for 5, 10, and 15 μg of proteins. The data are expressed as the protein band intensity value n ≥ 2, ∗, P ≤ 0.05 galactose versus glucose.
Nontransformed MRC5 fibroblasts grown in galactose for 15 doublings show an increased expression of some OXPHOS proteins. Expression levels of complex IV subunit 5a (COX 5a), complex I subunits 20 kDa (CI 20kD), porin, pyruvate dehydrogenase subunit E1α (PDH), complex III subunit (core 2), F1F0 ATPase subunit α (CV α), and complex II 70 kDa (CII 70kD) were examined by quantitative Western blot using monoclonal antibodies developed in our laboratory on glucose grown ( ) or galactose grown (
) or galactose grown ( ) MRC5 fibroblasts. The levels of the different complexes were determined by densitometry under conditions where the signal from bound antibodies was not saturated for 5, 10, and 15 μg of proteins. The data are expressed as the protein band intensity value n ≥ 2, ∗, P ≤ 0.05 galactose versus glucose.
) MRC5 fibroblasts. The levels of the different complexes were determined by densitometry under conditions where the signal from bound antibodies was not saturated for 5, 10, and 15 μg of proteins. The data are expressed as the protein band intensity value n ≥ 2, ∗, P ≤ 0.05 galactose versus glucose.
Glycolytic system remains high in galactose grown HeLa cells. Glucose using and glucose transport enzymes levels were examined by Western blot using antibodies against Hexokinase I, Hexokinase II, and Glut 1 in HeLa cells grown in glucose or galactose medium for 3 weeks. The levels of the different proteins obtained for glucose grown ( ) or galactose grown (
) or galactose grown ( ) HeLa cells were determined by densitometry under conditions where the signal from bound antibodies was not saturated (for 5, 10, and 15 μg of protein). No significant change was detected (n = 3).
) HeLa cells were determined by densitometry under conditions where the signal from bound antibodies was not saturated (for 5, 10, and 15 μg of protein). No significant change was detected (n = 3).
Glycolytic system remains high in galactose grown HeLa cells. Glucose using and glucose transport enzymes levels were examined by Western blot using antibodies against Hexokinase I, Hexokinase II, and Glut 1 in HeLa cells grown in glucose or galactose medium for 3 weeks. The levels of the different proteins obtained for glucose grown ( ) or galactose grown (
) or galactose grown ( ) HeLa cells were determined by densitometry under conditions where the signal from bound antibodies was not saturated (for 5, 10, and 15 μg of protein). No significant change was detected (n = 3).
) HeLa cells were determined by densitometry under conditions where the signal from bound antibodies was not saturated (for 5, 10, and 15 μg of protein). No significant change was detected (n = 3).
Mitochondrial DNA (mtDNA) content is unchanged in HeLa cells grown on galactose compared with glucose: mtDNA content was determined simultaneously with nuclear DNA content by real-time PCR in HeLa cells grown in glucose (A) or galactose (B). In A and B, the y axis represents the amplification of PCR products that are expressed as ΔRn, i.e., the amount of reporter signals detected during PCR amplification (no unit). mtDNA levels normalized relative to nuclear DNA signals (C) show no difference in mtDNA content/cell between HeLas grown in glucose or in galactose.
Mitochondrial DNA (mtDNA) content is unchanged in HeLa cells grown on galactose compared with glucose: mtDNA content was determined simultaneously with nuclear DNA content by real-time PCR in HeLa cells grown in glucose (A) or galactose (B). In A and B, the y axis represents the amplification of PCR products that are expressed as ΔRn, i.e., the amount of reporter signals detected during PCR amplification (no unit). mtDNA levels normalized relative to nuclear DNA signals (C) show no difference in mtDNA content/cell between HeLas grown in glucose or in galactose.
Mitochondria of HeLa cells and nontransformed MRC5 fibroblasts grown in galactose show the condensed configuration. The differences in mitochondrial structure between HeLa cells grown in glucose (A and B) or in galactose (D and E) were examined by electron microscopy. The same analysis was also performed in glucose-grown (C) or galatose-grown (F) MRC5 fibroblasts. Cells were fixed for Spurr’s resin embedment in 2.5% glutaraldehyde, 4% paraformaldehyde in 0.1 m sodium cacodylate (pH 7.4), with 8 mm CaCl2, followed by 1% osmium tetroxide (pH 7.4), followed by ethanol dehydration. Ultrathin sections were counterstained with lead citrate and viewed on a Philips CM12 electron microscope.
Mitochondria of HeLa cells and nontransformed MRC5 fibroblasts grown in galactose show the condensed configuration. The differences in mitochondrial structure between HeLa cells grown in glucose (A and B) or in galactose (D and E) were examined by electron microscopy. The same analysis was also performed in glucose-grown (C) or galatose-grown (F) MRC5 fibroblasts. Cells were fixed for Spurr’s resin embedment in 2.5% glutaraldehyde, 4% paraformaldehyde in 0.1 m sodium cacodylate (pH 7.4), with 8 mm CaCl2, followed by 1% osmium tetroxide (pH 7.4), followed by ethanol dehydration. Ultrathin sections were counterstained with lead citrate and viewed on a Philips CM12 electron microscope.
Mitochondrial reticulum thins in galactose grown HeLa cells: the overall morphology of the entire mitochondrial network was studied by confocal microscopy (Nikon eclipse E600FN-Bio-Rad radiance 2100 MP) in HeLa cells (A and C) or MRC5 fibroblasts (B and D) transfected with a mitochondrial-targeted green fluorescent protein and grown for 15 doublings in either glucose (A and B) or galactose medium (C and D). Note the thinning and the higher networking of the mitochondrion in HeLas grown in galactose (C). These pictures represent the differences resulting from several observations realized on living cells using a dip in lens (60X Fluor 1.00W-Nikon) in a Petri dish containing growth medium maintained at 37°C.
Mitochondrial reticulum thins in galactose grown HeLa cells: the overall morphology of the entire mitochondrial network was studied by confocal microscopy (Nikon eclipse E600FN-Bio-Rad radiance 2100 MP) in HeLa cells (A and C) or MRC5 fibroblasts (B and D) transfected with a mitochondrial-targeted green fluorescent protein and grown for 15 doublings in either glucose (A and B) or galactose medium (C and D). Note the thinning and the higher networking of the mitochondrion in HeLas grown in galactose (C). These pictures represent the differences resulting from several observations realized on living cells using a dip in lens (60X Fluor 1.00W-Nikon) in a Petri dish containing growth medium maintained at 37°C.
Live cell functional measurements demonstrate a more active state of oxidative phosphorylation in cells grown in galactose medium compared with glucose
| Functional measurements . | Hela (304I) . | . | |
|---|---|---|---|
| . | Glucose . | Galactose . | |
| Doubling timea | 18.4 ± 01 | 58 ± 02 | |
| Respiratory rateb | 1.35 ± 0.23 | 2.72 ± 0.46 | |
| Cytochrome c oxydase activityc | 5.6 ± 1.38 | 17.35 ± 5.04 | |
| Matrix pHd | 8.03 ± 0.05 | 7.65 ± 0.10 | |
| Matrix pH after CCCPe | 6.9 ± 0.12 | 6.82 ± 0.19 | |
| Matrix redox statef | 93 ± 3.0 | 74 ± 2.5 | |
| Functional measurements . | Hela (304I) . | . | |
|---|---|---|---|
| . | Glucose . | Galactose . | |
| Doubling timea | 18.4 ± 01 | 58 ± 02 | |
| Respiratory rateb | 1.35 ± 0.23 | 2.72 ± 0.46 | |
| Cytochrome c oxydase activityc | 5.6 ± 1.38 | 17.35 ± 5.04 | |
| Matrix pHd | 8.03 ± 0.05 | 7.65 ± 0.10 | |
| Matrix pH after CCCPe | 6.9 ± 0.12 | 6.82 ± 0.19 | |
| Matrix redox statef | 93 ± 3.0 | 74 ± 2.5 | |
The doubling time (DT) was calculated according to the formula: DT = 0.693.t/1n (N/N0) with t (elapsed time), N0 (starting number of cells), N (final number of cells). DT is expressed in hours (n = 2 different experiments).
Expressed in nmol O2/min/106 cells (n ≥ 4).
Expressed in nmol O2/min/106 cells (n ≥ 4).
n ≥ 10.
n ≥ 10.
Expressed as percentage reduction of rosGFP1 (n ≥ 3).
Substrate-dependent changes in mitochondrial morphology
| Morphometric measurements . | Hela (304I) . | . | |
|---|---|---|---|
| . | Glucose . | Galactose . | |
| Mitochondria/Cella | 5.4 ± 0.94 | 4.2 ± 1.96 | |
| Mitochondrial profiles/Cellb | 15.6 ± 6.8 | 19.8 ± 7.3 | |
| Mitochondrial section areac | 0.31 ± 0.22 | 0.22 ± 0.18 | |
| Mitochondrial tubular diameterd | 0.48 ± 0.08 | 0.32 ± 0.07 | |
| Intracristal/mitochondrial areae | 8 ± 1 | 24 ± 3 | |
| Matrix densityf | 11.6 ± 3 | 45.6 ± 8 | |
| Cristal/inner membraneg | 1.5 ± 0.66 | 3.6 ± 1.6 | |
| Number of inclusion bodiesh | 1 | 25 | |
| Morphometric measurements . | Hela (304I) . | . | |
|---|---|---|---|
| . | Glucose . | Galactose . | |
| Mitochondria/Cella | 5.4 ± 0.94 | 4.2 ± 1.96 | |
| Mitochondrial profiles/Cellb | 15.6 ± 6.8 | 19.8 ± 7.3 | |
| Mitochondrial section areac | 0.31 ± 0.22 | 0.22 ± 0.18 | |
| Mitochondrial tubular diameterd | 0.48 ± 0.08 | 0.32 ± 0.07 | |
| Intracristal/mitochondrial areae | 8 ± 1 | 24 ± 3 | |
| Matrix densityf | 11.6 ± 3 | 45.6 ± 8 | |
| Cristal/inner membraneg | 1.5 ± 0.66 | 3.6 ± 1.6 | |
| Number of inclusion bodiesh | 1 | 25 | |
Total mitochondrial area expressed as percentage of total cell area transmission electron microscopy (TEM) n ≥ 100.
Average number of mitochondrial profiles within cell section (TEM) n ≥ 100.
Average area of a mitochondrial cross-section (TEM) n ≥ 100.
Average diameter of the mitochondrial tubule (fluorescence), n ≥ 100.
Total cristal area expressed as percentage of total mitochondrial area, n ≥ 100.
Average density of matrix (TEM) n ≥ 100.
Total cristal membrane divided by inner membrane n ≥ 100.
Number of inclusion bodies observed n ≥ 100.
Acknowledgments
We thank Kathy Chicas-Cruz for helping with the cell culture and Bruce Birrell and Andy Marcus for sharing their equipment. We also thank Jeanne Selker for fixation and embedding of EM samples and Beth Prescott and Mike Marusich for helpful advice on cell culture.